
 |
Manual |
The enhanced van Holde - Weischet analysis is a model independent method to analyze velocity data. It will provide S-value distributions that are corrected for diffusion effects, and provide valuable diagnostics for velocity data. Compared to the traditional van Holde - Weischet method there are several improvements in the enhanced method which allow it to better deal with heterogeneous samples and samples that contain two or more well-separated components. The enhanced method also can take advantage of data points near the bottom of the cell and from scans that have no longer a stable plateau or have not yet cleared the meniscus.
You start the enhanced van Holde - Weischet data analysis by clicking on "Enhanced van Holde - Weischet" in the Velocity sub-menu of the main menu. The main Enhanced van Holde - Weischet data analysis window will appear.
The first step in the analysis requires that you load an UltraScan dataset that has previously been edited with the UltraScan editing module. UltraScan sedimentation velocity datasets always have the suffix ".us.v". Click on the "Load Dataset" button in the left upper corner of the control panel. Simply select the desired run from the selection of available runs in the dataset loading dialog. Once loaded, the Run Details will be shown. Click on the "Accept" or "Cancel" button, and you will be returned to the analysis window, which will show the first available dataset of the selected run in the edited data window on the lower right panel, and the enhanced van Holde - Weischet analysis will be shown in the upper right panel of the analysis window.
Analysis Functions:
 |
Click on these buttons to control the enhanced van Holde - Weischet analysis.
|
Run Information:
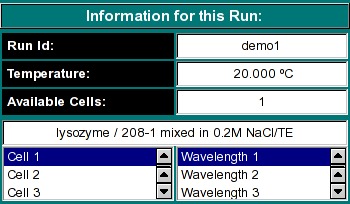 |
|
Experimental Parameters:
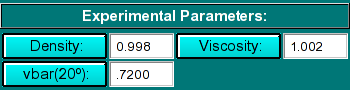 |
Here you can enter the corrections for density, viscosity, and partial
specific volume of your sample. As you change the information in these
fields, the program will automatically update the analysis to correct the
S-value according to the specified buffer conditions. If the data was
retrieved from the database and associated with the correct buffer file
and peptide file, the density, viscosity and partial specific volume (vbar)
will automatically be updated to the appropriate values and don't need to be
adjusted to produce S 20,W corrected values.
|
Analysis Controls:
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on July 17, 2005.