

|
Manual |
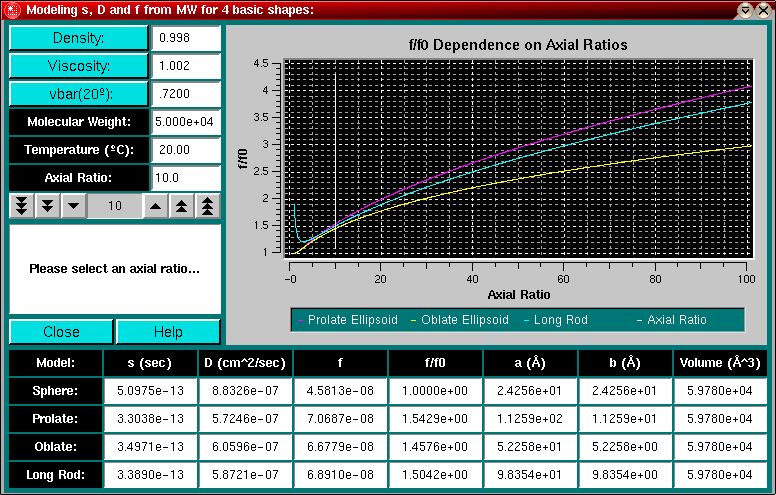
You can start this model from the Simulation Menu entry in the Main Menu. Given a molecular weight, partial specific volume, and an axial ratio, this module allows you to model the molecular dimensions of a sphere, prolate ellipsoid, oblate ellipsoid and a long rod. Further, by providing the buffer conditions and temperature observed in a sedimentation run, this program will calculate the sedimentation, diffusion, and frictional coefficient for these hypothetical models.
The program is useful for the generation of simulation input parameters for the finite element simulation routine. If you have some idea about the possible size, overall shape and the run conditions of the planned experiment, you could first simulate the hydrodynamic parameters and then enter them into the finite element simulation program. With the finite element simulation program you could then try out different speeds to find the optimal conditions under which to perform your experiment.
Use the Density and Viscosity buttons to find the appropriate values from the buffer tables or enter the desired values directly into the respective text fields. Use the Vbar (20oC) button to estimate the partial specific volume from a protein sequence, or enter the partial specific volume, corrected for 20oC, directly into the corresponding text field. Indicate the expected molecular weight of your sample and set the temperature to the run temperature. Select an axial ratio by using the counter or by entering the ratio into the text field.
Each time a parameter is changed, the plot area and affected text fields are updated.
This document is part of the UltraScan Software Documentation
distribution.
Copyright © notice.
The latest version of this document can always be found at:
Last modified on January 12, 2003.